| Kidney Res Clin Pract > Volume 42(4); 2023 > Article |
|
Abstract
Background
Methods
Results
Acknowledgments
Notes
Funding
This work was supported by a National Research Foundation of Korea (NRF) grant funded by the Korean government (Ministry of Science and ICT, MSIT) (No. 2021R1A2C2094586). This study was supported by the 2022 Young Investigator Research Grant from the Korean Nephrology Research Foundation. The study was performed independently by the authors.
Data sharing statement
The data used for this study are available in the public database. The CKDGen data are openly available on the Consortium website (https://ckdgen.imbi.uni-freiburg.de/), and the UK Biobank data is accessible after acquiring approval from the organization (application No. 53799).
Authors’ contributions
Conceptualization, Formal analysis: SP, DKK
Data curation: SP, SGK, SL, YCK, SC, KK
Funding acquisition: YCK, SSH, HL, JPL, KWJ, CSL, YSK, DKK
Investigation: SP, SGK, SL, YCK, SC
Methodology: SP, KK, DKK
Project administration: SGK, SL, YK, SC
Resources, Supervision: KK, YCK, SSH, HL, JPL, KWJ, CSL, YSK, DKK
Software: KK, DKK
Validation, Visualization: SP, SGK, SL
Writing–original draft: SP, SGK, SL, YK, SC, KK
Writing–review & editing: All authors
All authors read and approved the final manuscript.
Supplementary Materials
Figure 1.
Mendelian randomization analysis scatter plot.
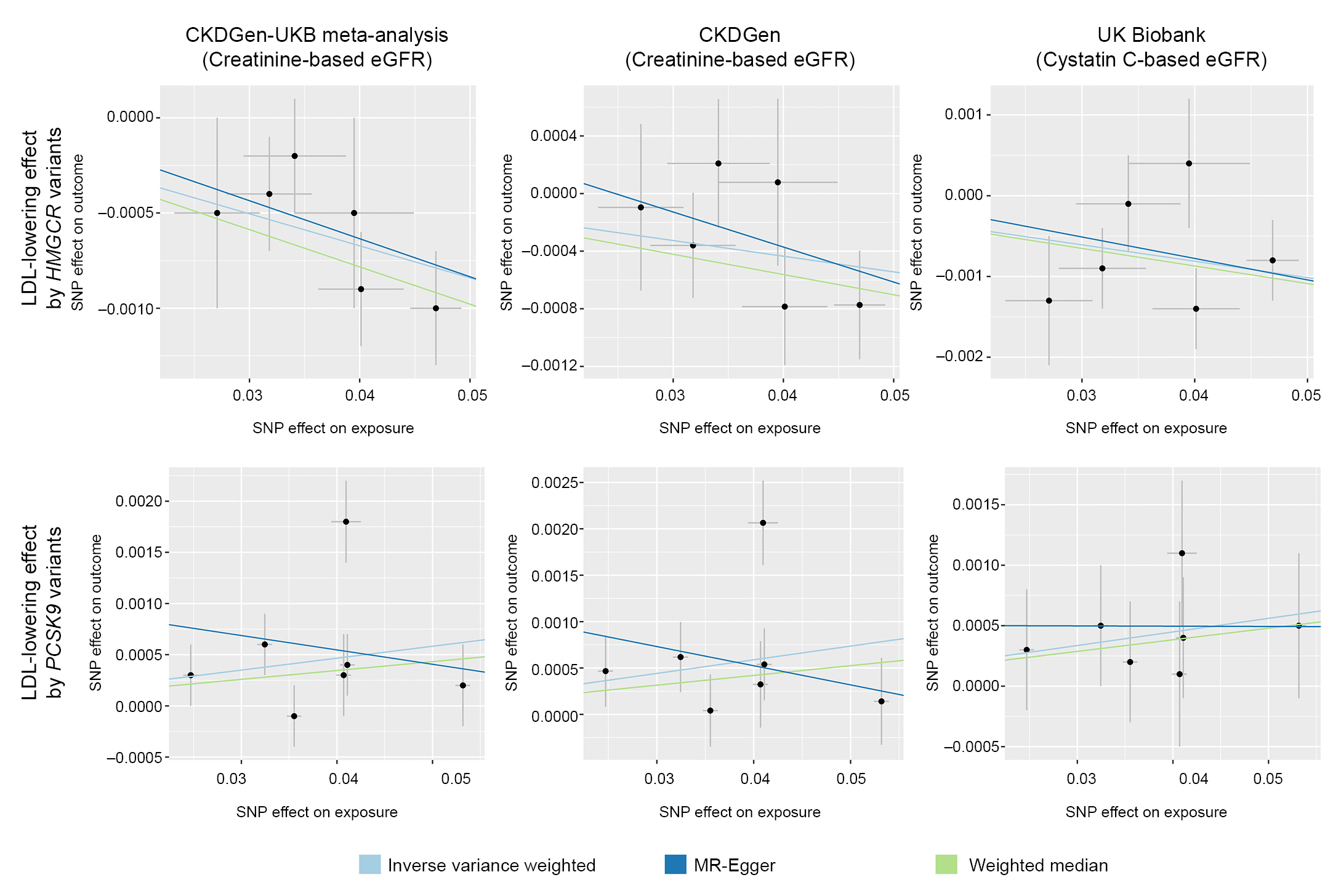
Figure 2.
Single-SNP analysis results.
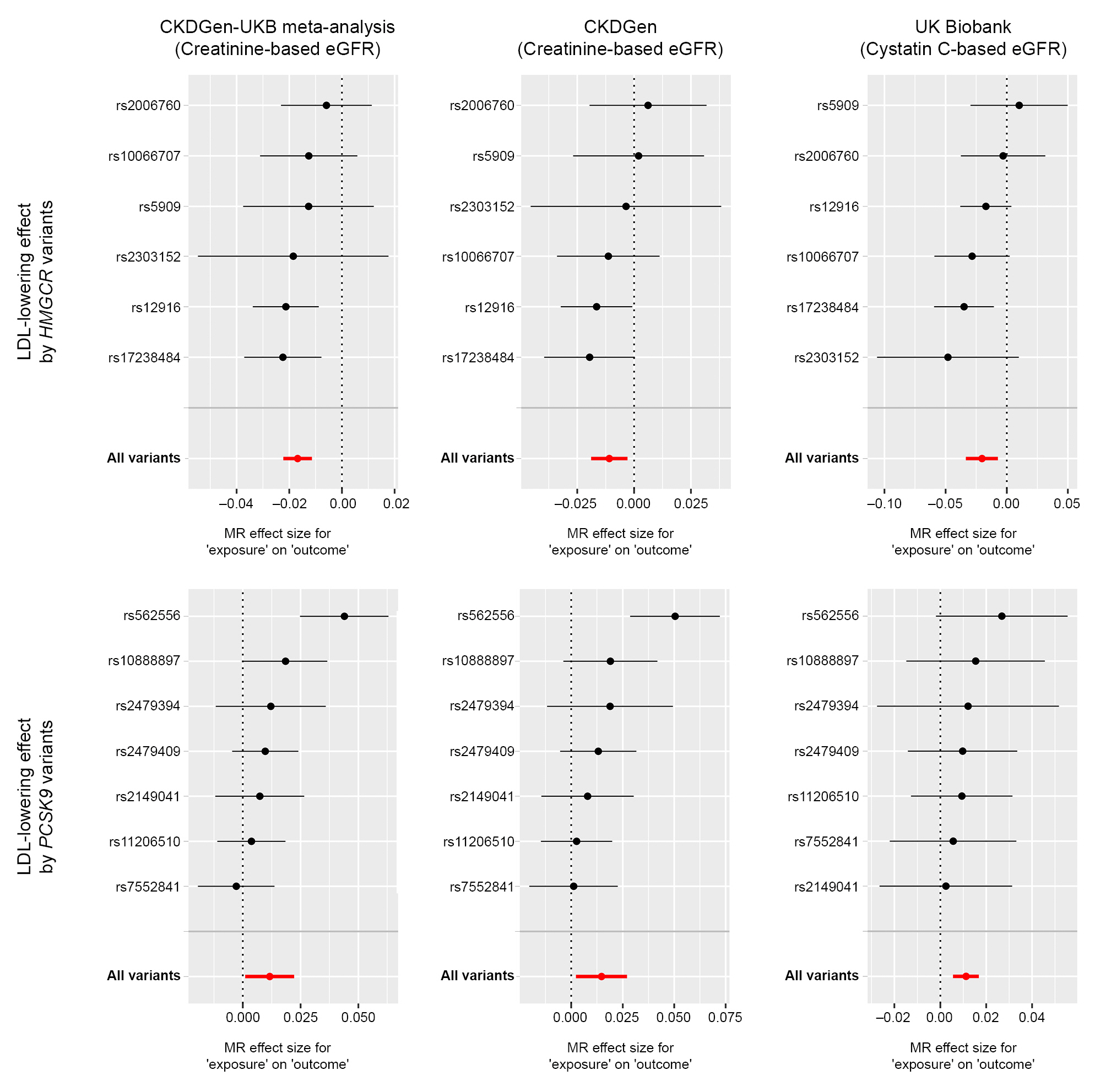
Figure 3.
Leave-one-out analysis results.
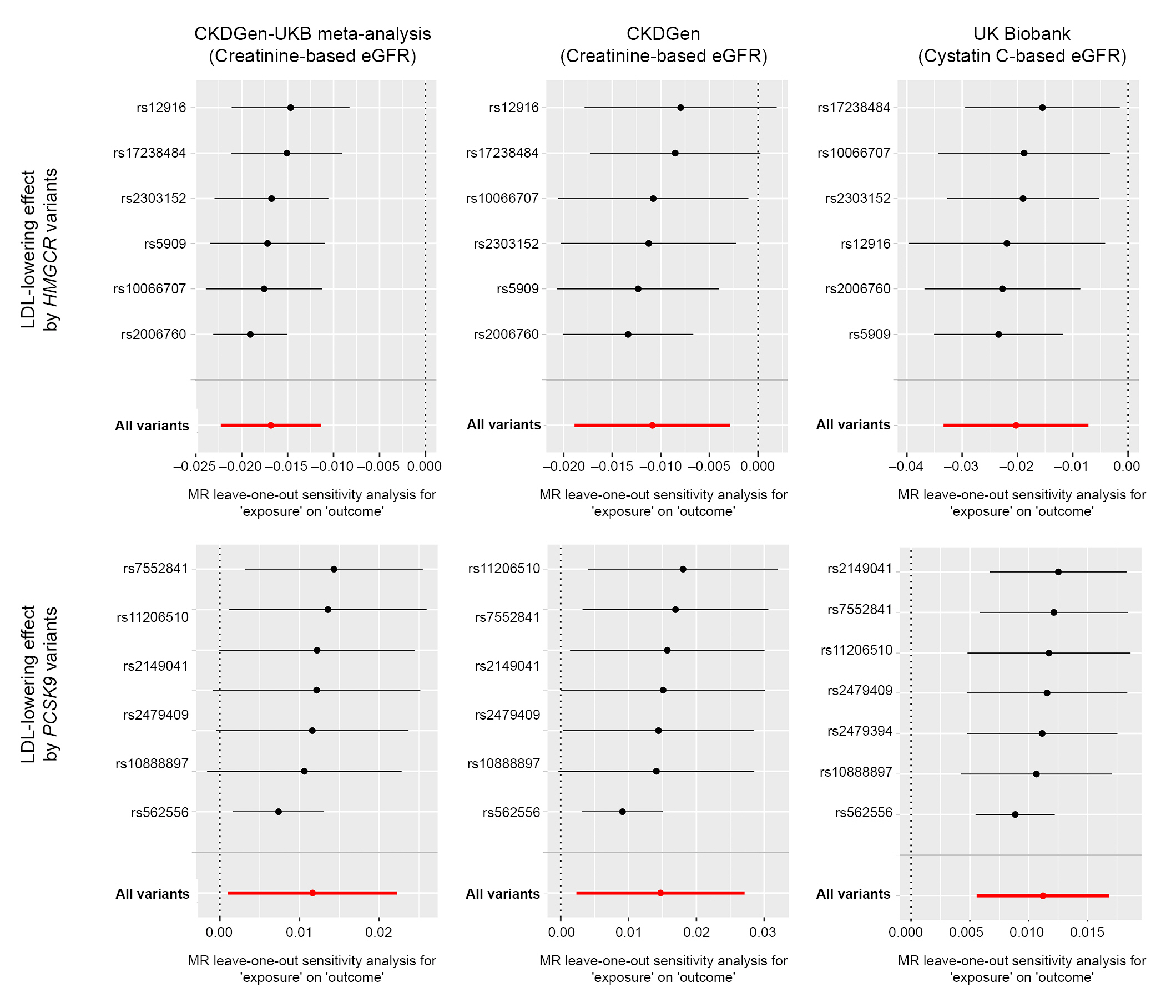
Figure 4.
Nonlinear MR analysis results.

Table 1.
All effect sizes were aligned and scaled toward a genetically predicted 50-mg/dL decrease in LDL cholesterol by instrumented variants.
eGFR, estimated glomerular filtration rate; LDL, low-density lipoprotein cholesterol; MR, Mendelian randomization; MR-IVW, multi-replicative inverse variance weighted; UKB, UK Biobank.
References
- TOOLS
-
METRICS

- ORCID iDs
-
Sehoon Park

https://orcid.org/0000-0002-4221-2453Seong Geun Kim

https://orcid.org/0000-0002-8831-9838Soojin Lee

https://orcid.org/0000-0001-5633-3961Yaerim Kim

https://orcid.org/0000-0003-1596-1528Kwangsoo Kim

https://orcid.org/0000-0002-4586-5062Yong Chul Kim

https://orcid.org/0000-0003-3215-8681Seung Seok Han

https://orcid.org/0000-0003-0137-5261Hajeong Lee

https://orcid.org/0000-0002-1873-1587Jung Pyo Lee

https://orcid.org/0000-0002-4714-1260Kwon Wook Joo

https://orcid.org/0000-0001-9941-7858Chun Soo Lim

https://orcid.org/0000-0001-9123-6542Yon Su Kim

https://orcid.org/0000-0003-3091-2388Dong Ki Kim

https://orcid.org/0000-0002-5195-7852 - Related articles



 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Download Citation
Download Citation Supplement 1
Supplement 1 Print
Print